SEPALLATA (SEP) genes are the plant-specific MIKC-type MADS-box genes, belonging to the class E genes in the ABCE model of flower organ development. The function of SEPs in the regulation of floral organ development is widely present in plants. However, the function of JcSEP genes in regulating flower development remains unclear in Jatropha curcas, a promising biofuel plant.
In a study published in Planta, researchers from Xishuangbanna Tropical Botanical Garden (XTBG) investigated the role of the JcSEP3 gene in floral organ development in the woody plant Jatropha curcas.
To investigate the function of JcSEP3, the researchers constructed the JcSEP3-overexpressing and JcSEP3-RNA interference (RNAi) vectors to transform Jatropha. Positive transgenic plants were further confirmed using qRT-PCR analysis.
They found that the overexpressing JcSEP3 in inflorescence buds caused abnormal phenotypes of the JcSEP3-overexpressing transgenic plants, which implies that JcSEP3 is an important regulator in the development of stamens of Jatropha.
To further understand genetic changes in transgenic plants at genome-wide level, the researchers compared transcriptome changes in inflorescences between the JcSEP3-overexpressing, JcSEP3-RNAi transgenic and wild-type plants.
In the JcSEP3-overexpressing transgenic plants, they discovered 25 upregulated genes involved in anther and pollen development, as well as 12 induced genes in brassinosteroid (BR) and gibberellin (GA) signaling pathways.
“Our results suggest that JcSEP3 directly or indirectly regulates stamen development, concomitant with the regulation of BR and GA signaling pathways,” said CHEN Maosheng of XTBG.
Contact
CHEN Maosheng Ph.D
Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun 666303, Yunnan, China
E-mail: chenms@xtbg.org.cn
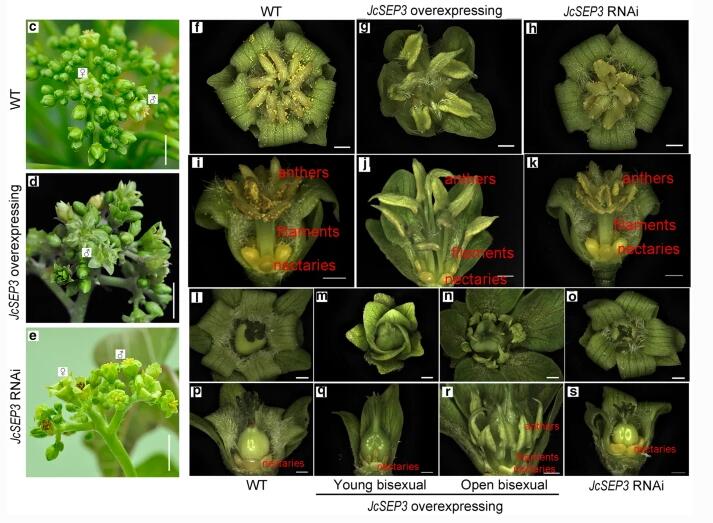
Phenotypes of wild-type (WT), JcSEP3-overexpressing and JcSEP3-RNAi transgenic plants. (Image by ZHAO Meili)

